New paper: Geography and genetics in Salamandra algira - historical influences and contemporary patterns
A new paper, published with a wonderful large set of collaborators, resolves the influence of postglacial dynamics and climatic oscillations on the genetic diversity of the Algerian salamander. This species is highly variable in colouration and some populations are of conservation concern.
Dinis M, Merabet K, Martinez-Freiria F, Steinfartz S, Vences M, Burgon JD, Elmer KR, Donaire D, Hinckley A, Fahd S, Joger U, Fawzi A, Slimani T, Velo-Anton G (2018) Allopatric diversification and evolutionary melting pot in a North African Palearctic relict: The biogeographic history of Salamandra algira. Molecular Phylogenetics and Evolution, in press,
The paper is currently freely available here:
https://authors.elsevier.com/c/1XvLg3m3nMuJY1
And then soon will be fully open access, thanks to RCUK and Uni Glasgow.
Abstract
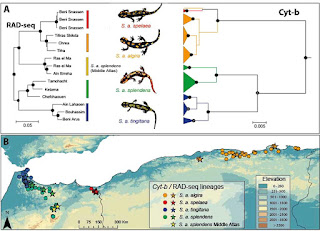
North Africa is a climatically and topographically complex region with unique biotic assemblages resulting from the combination of multiple biogeographic realms. Here, we assess the role of climate in promoting intra-specific diversification in a Palearctic relict, the North African fire salamander, Salamandra algira, using a combination of phylogenetic and population genetic analyses, paleoclimatic modelling and niche overlap tests. We used mitochondrial DNA (Cyt-b), 9838 ddRADseq loci, and 14 microsatellite loci to characterize patterns of genetic diversity and population structure. Phylogenetic analyses recover two major clades, each including several lineages with mito-nuclear discordances suggesting introgressive patterns between lineages in the Middle Atlas, associated with a melting pot of genetic diversity. Paleoclimatic modelling identified putative climatic refugia, largely matching areas of high genetic diversity, and supports the role of aridity in promoting allopatric diversification associated with ecological niche conservatism. Overall, our results highlight the role of climatic microrefugia as drivers of populations’ persistence and diversification in the face of climatic oscillations in North Africa, and stress the importance of accounting for different genomic regions when reconstructing biogeographic processes from molecular markers.
Dinis M, Merabet K, Martinez-Freiria F, Steinfartz S, Vences M, Burgon JD, Elmer KR, Donaire D, Hinckley A, Fahd S, Joger U, Fawzi A, Slimani T, Velo-Anton G (2018) Allopatric diversification and evolutionary melting pot in a North African Palearctic relict: The biogeographic history of Salamandra algira. Molecular Phylogenetics and Evolution, in press,
The paper is currently freely available here:
https://authors.elsevier.com/c/1XvLg3m3nMuJY1
And then soon will be fully open access, thanks to RCUK and Uni Glasgow.
Abstract
North Africa is a climatically and topographically complex region with unique biotic assemblages resulting from the combination of multiple biogeographic realms. Here, we assess the role of climate in promoting intra-specific diversification in a Palearctic relict, the North African fire salamander, Salamandra algira, using a combination of phylogenetic and population genetic analyses, paleoclimatic modelling and niche overlap tests. We used mitochondrial DNA (Cyt-b), 9838 ddRADseq loci, and 14 microsatellite loci to characterize patterns of genetic diversity and population structure. Phylogenetic analyses recover two major clades, each including several lineages with mito-nuclear discordances suggesting introgressive patterns between lineages in the Middle Atlas, associated with a melting pot of genetic diversity. Paleoclimatic modelling identified putative climatic refugia, largely matching areas of high genetic diversity, and supports the role of aridity in promoting allopatric diversification associated with ecological niche conservatism. Overall, our results highlight the role of climatic microrefugia as drivers of populations’ persistence and diversification in the face of climatic oscillations in North Africa, and stress the importance of accounting for different genomic regions when reconstructing biogeographic processes from molecular markers.